

In mice, intraperitoneal administration of JD1 reduced tissue colonization by S. Cholesterol, a component of mammalian cell membranes, was protective in the presence of neutral lipids. coli inner membranes versus mammalian cell membranes. Moreover, JD1 preferentially damaged liposomes with compositions similar to E. We quantified macrophage cell membrane integrity and mitochondrial membrane potential and found that disruption of eukaryotic cell membranes required approximately 30-fold more JD1 than was needed to kill bacteria in macrophages. Using a combination of cellular indicators and super resolution microscopy, we found that JD1 damaged bacterial cytoplasmic membranes by increasing fluidity, disrupting barrier function, and causing the formation of membrane distortions.
JD1 is not antibacterial in standard microbiological media, but rapidly inhibits growth and curtails bacterial survival under broth conditions that compromise the outer membrane or reduce efflux pump activity. Typhimurium) residing within macrophages. Using chemical genetics, we recently identified a small molecule, JD1, that kills Salmonella enterica serovar Typhimurium ( S. However, mammals have evolved a substantial immune arsenal that weakens pathogen defenses, suggesting the feasibility of developing therapies that work in concert with innate immunity to kill Gram-negative bacteria. Biophysical Journal, 93:496-504, 2007.įeatured in Science Magazine’s Editors’ Choice: Science, 316:1101, 2007.Infections caused by Gram-negative bacteria are difficult to fight because these pathogens exclude or expel many clinical antibiotics and host defense molecules.
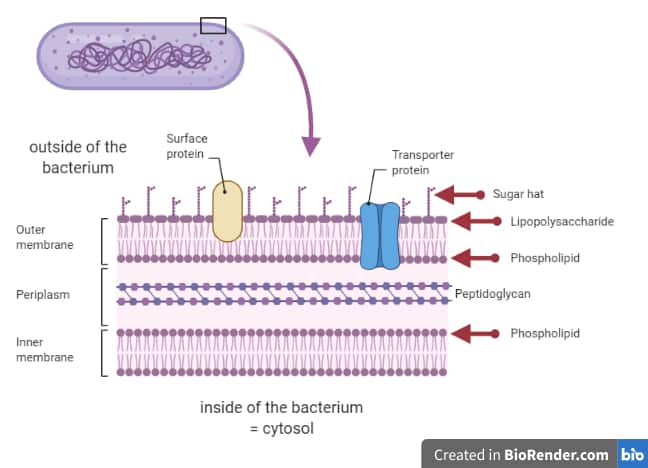
Mechanics of force propagation in TonB-dependent outer membrane transport. Coupling of calcium and substrate binding through loop alignment in the outer membrane transporter BtuB. Molecular basis for activation of a catalytic asparagine residue in a self-cleaving bacterial autotransporter. Steere, O. Zak, P. Aisen, E. Tajkhorshid, R. W. Structural basis for iron piracy by pathogenic Neisseria. This conformation is predicted to greatly accelerate the cleavage reaction, and the other residues found to be involved are in agreement with experimental observations. The simulations reveal an unusual conformation of the catalytic asparagine residue, seen in less than 2% of all atomic structures, that is supported by the unique electrostatic environment created by neighboring residues. We have carried out simulations on the native pre-cleaved state as well as on a number of biochemically characterized mutants. After transport, the N-terminal domain is typically self-cleaved, freeing it from the membrane. For example, certain virulence factors are secreted through the use of so-called “auto-transporters”, proteins that export themselves! The auto-transporter consists of two domains, a membrane-bound barrel at the C-terminus and a secreted domain at the N-terminus. Just as bacteria need to import molecules across the outer membrane, they sometimes need to export them as well. Model of the full-length autotransporter EspP. In particular, we see in multiple contexts that the interior domain of the transporter unfolds, generating a pathway for the substrate across the membrane. We have proposed that energy is transduced through a mechanical coupling between the inner and outer membrane complexes, supported by simulations that demonstrate the strength of the TonB-transporter connection mediated by hydrogen bonds, as well as the response of the transporter to the application of force. The mechanisms of this interaction and how it causes transport are still open questions in the field of microbiology. To get energy for this task, they utilize an inner membrane protein, TonB, that couples across the periplasm to an outer membrane transporter for import of, e.g., vitamin B12 or iron extracted from human transferrin. Nonetheless, bacteria often need to actively import large and/or scarce nutrients across the outer membrane. As such, energy cannot be generated or stored here. GRAM NEGATIVE BACTERIA MEMBRANE FULL
TonB (red) “pulling” on the lumenal domain of the transporter BtuB (blue), the connection being mediated by hydrogen bonds (yellow).Īnother unique aspect of the outer membrane is that it’s full of porins, which permit the diffusion of small molecules relatively easily across it.






 0 kommentar(er)
0 kommentar(er)
